특징
- CRISPR interference (CRISPRi)-Ready ioGlutamatergic Neuron은 inactive Cas9 nuclease (dCas9)을 발현하도록 설계된 iPSC neuron 입니다.
- 해동 후 1일째부터 guide RNA (gRNA)를 도입함으로써, dCas9이 최소 21일 동안 높은 수준으로 발현됩니다.
- 최적화된 lentivirus gRNA 전달을 통해 유전자 억제, CRISPR 억제 스크리닝을 수행하고 며칠 이내에 실험 결과를 측정할 수 있습니다.
- CRISPRi-Ready, CRISPRa-Ready, CRISPRko-Ready ioGlutamatergic neuron을 사용하면 Cas9 안정형 iPSC line을 엔지니어링하고 특성화하거나 분화 프로토콜을 최적화하는데에 있어 실험 일정이 크게 단축됩니다.
사양
| Starting material |
Human iPSC line |
| Donor |
Caucasian adult male, age 55-60 years old
(skin fibroblast) |
| Format |
Cryopreserved cells |
| Differentiation
method |
opti-ox deterministic programming |
| Karyotype |
Normal (46, XY) |
| Vial size |
Small: >1 x 10⁶ viable cells |
| Recommended seeding
density |
30,000 cells/cm² |
| Seeding
compatibility |
6, 24, 96 well plates |
| Quality control |
Sterility, protein expression (ICC), gene
expression (RT-qPCR), functionality of CRISPRi (flow cytometry) |
| Application |
Gene
repressions
Pooled CRISPR interference screens
Arrayed CRISPR interference screens |
프로토콜
냉동 보존 형식으로 제공되며 권장 배지에서 재생 시 빠르게 성숙되도록 프로그래밍되어 있습니다.
Phase 0 단계의 세포를 수령 후, Phase 1, 2 과정을 거쳐 실험에 바로 이용합니다.
1) Phase 0 : Induction (bit.bio에서 Induction 후 제공)
2) Phase 1 : 고객이 세포를 수령 후 plating 하여 안정화시키는 기간 (4일)
3) Phase 2 : 세포의 성숙을 유지하는 기간
* guide RNA 전달은 회복 후 1-11일 사이에 수행 / guide RNA 전달 후 5일째 부터 read out 가능

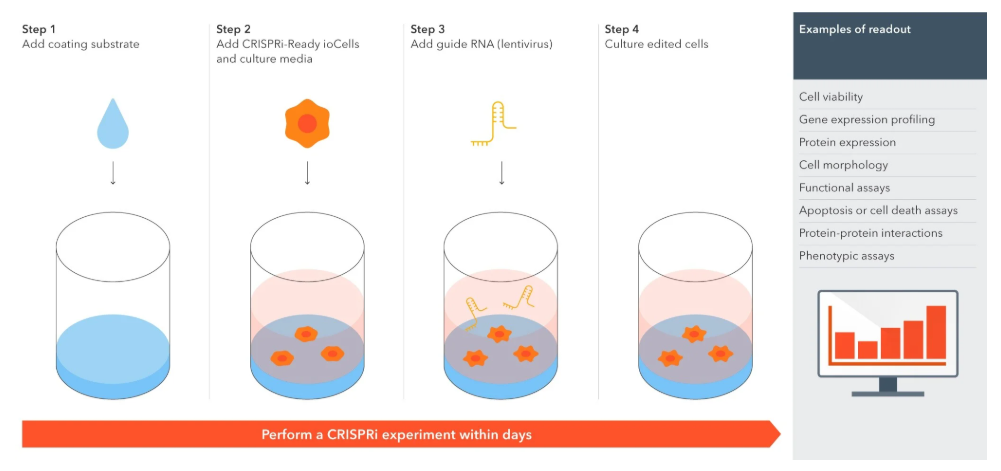
실험 과정
참고 데이터
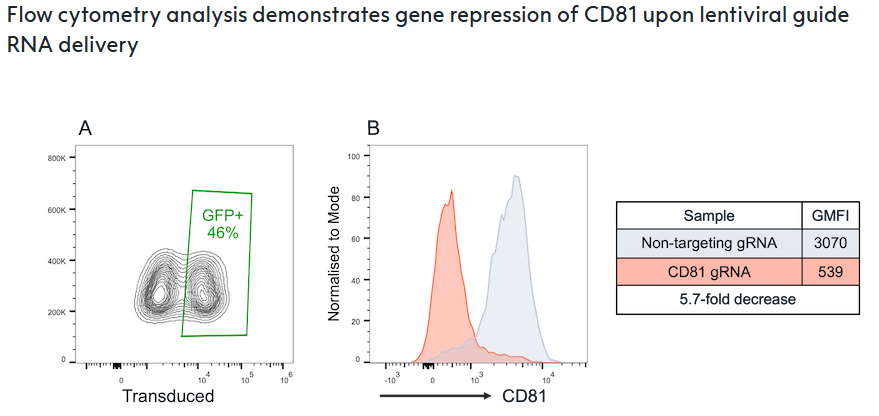
Flow cytometry analysis confirms robust CD81 gene repression in CRISPRi-Ready ioGlutamatergic Neurons following lentiviral delivery of a CD81-targeting gRNA on day 3 post-thaw. Gene repression was measured by flow cytometry after five days of culture.
(A) 45.8% of cells received the CD81-targeting gRNA via lentiviral transduction, as indicated by GFP expression.
(B) Functionality of the dCas9-based transcriptional repressor is demonstrated by a 5.7-fold decrease in CD81 protein expression (red histogram) compared to non-targeting gRNA controls (grey histogram), measured by geometric mean fluorescence intensity (GMFI) in the GFP+ population.
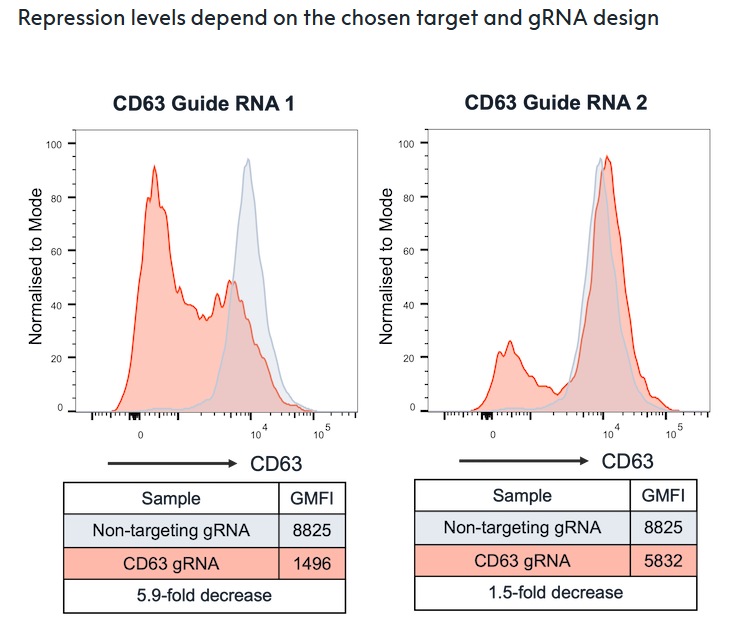
Flow cytometry analysis of CD63 protein expression in CRISPRi-Ready ioGlutamatergic Neurons 5 days after delivery of two different gRNAs. Guide RNAs were delivered on day 3 post-thaw via lentiviral transduction.
Designing gRNAs for CRISPR interference is complex and requires precise targeting of regulatory regions near the transcription start site; efficacy is significantly affected by factors such as chromatin accessibility and epigenetic modifications.
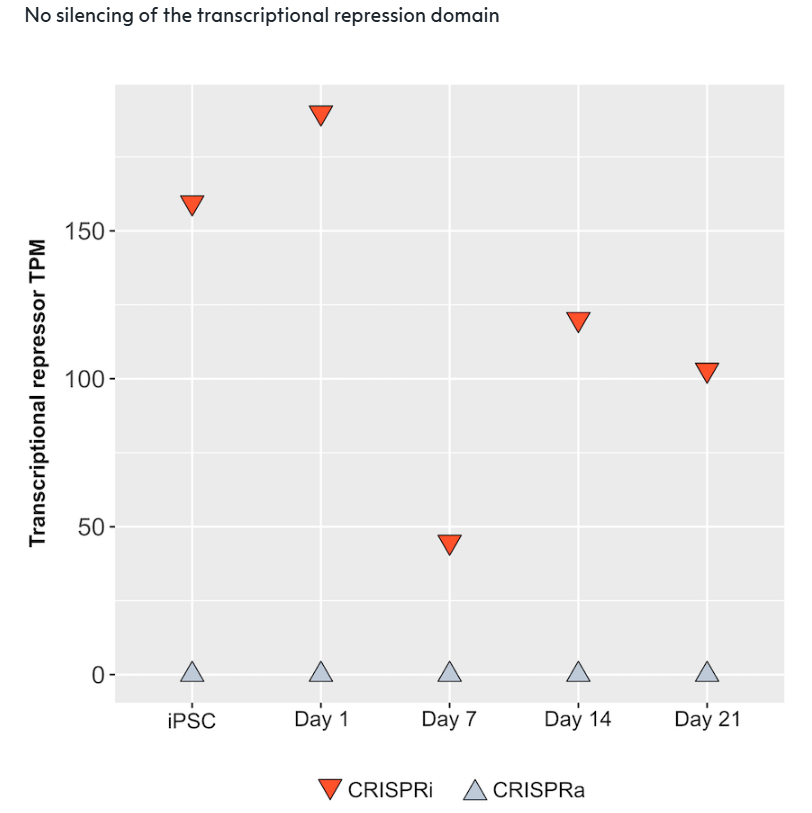
Bulk RNA sequencing analysis was performed on CRISPRi-Ready ioGlutamatergic Neurons (CRISPRi) and CRISPRa-Ready ioGlutamatergic Neurons (CRISPRa) at the iPSC stage and on days 1, 7, 14 and 21 post-revival.
Gene expression profiling revealed sustained transcriptional repressor expression in CRISPRi-Ready ioGlutamatergic Neurons throughout the culture period, while no expression of the transcriptional repressor was detected in the control line (CRISPRa-Ready ioGlutamatergic Neurons).