특징
- Cas9 nuclease를 지속적으로 발현하도록 설계되었습니다.
- 해동 후 1-18일 사이에 guide RNA (gRNA)를 바로 도입할 수 있습니다.
- Bit.Bio에서 제공하는 최적화된 렌티바이러스 또는 lipid 기반 gRNA 도입 프로토콜을 사용하면, 녹아웃 효율이 극대화되고, 며칠 이내에 유전자 녹아웃 및 CRISPR 판독값을 측정할 수 있습니다.
- 수일 이내에 P2RY12 및 IBA1을 90% 이상 발현하는 microglia로 전환됩니다.
- 사용자가 자신의 Cas9 stable iPSC line을 엔지니어링하거나 특성화하거나 분화프로토콜을 최적화 하는데 들어가는 수개월의 시간을 전약할 수 있습니다. 단지 gRNA만 도입하여 견고한 실험 결과를 바로 얻을 수 있습니다.
사양
| Starting
material |
Human iPSC line |
| Donor |
Caucasian adult male (skin
fibroblast) |
| Format |
Cryopreserved cells |
| Differentiation
method |
opti-ox deterministic programming |
| Karyotype |
Normal (46, XY) |
| Vial size |
Small: >1.5 x 10⁶ viable cells |
| Recommended
seeding density |
37,000 to 39,500 cells/cm² |
| Seeding
compatibility |
6, 12, 24, 48, 96 & 384 well
plates |
| Quality
control |
Sterility,
protein expression (ICC), functional phagocytosis and cytokine secretion
assays,
Cas9 functional validation (flow cytometry) |
| Applications |
Single gene
knockouts
Combinatorial gene knockouts
Pooled CRISPR screens
Arrayed CRISPR screens
High throughput screening |
프로토콜
냉동 보존 형식으로 제공되며 권장 배지에서 재생 시 빠르게 성숙되도록 프로그래밍되어 있습니다.
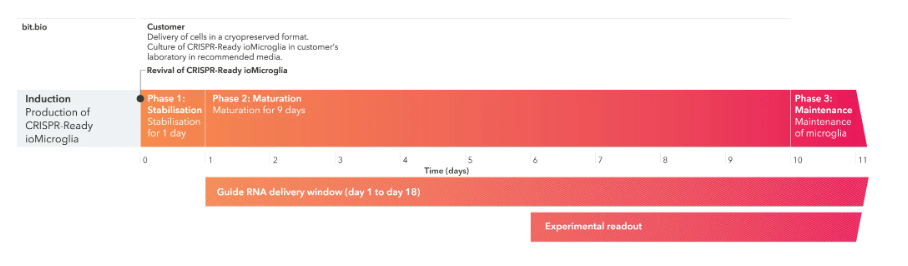
Phase 0 단계의 세포를 수령 후, Phase 1, 2,3 과정을 거쳐 실험에 바로 이용합니다.
1) Phase 0 : Induction (bit.bio에서 Induction 후 제공)
2) Phase 1 : 고객이 세포를 수령 후 plating 하여 안정화시키는 기간 (1일)
3) Phase 2 : 세포가 성숙하는 기간 (9일)
4) Phase 3 : 유지 기간 (1일)
* gRNA는 해동 후 1-18일 사이에 도입될 수 있으며, 도입 후 5일 후에 판독을 수행할 수 있습니다.
실험 과정
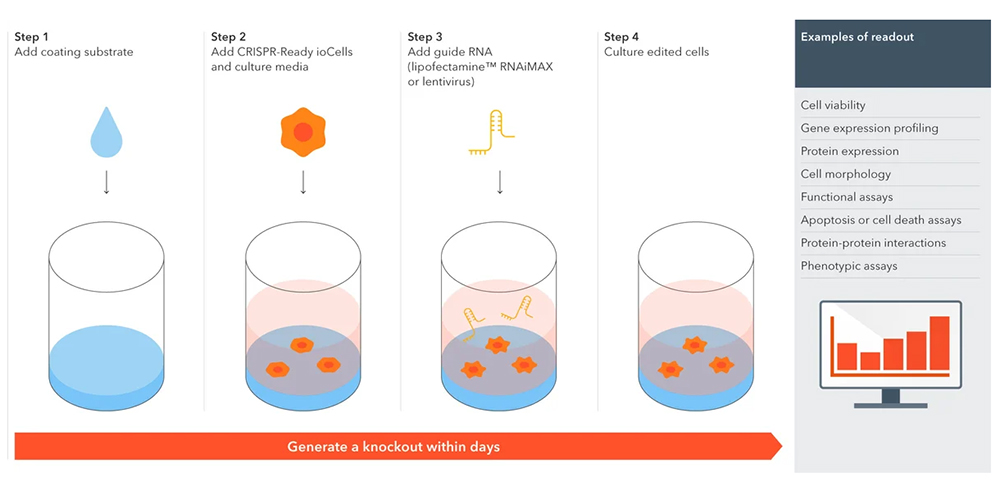
참고 데이터
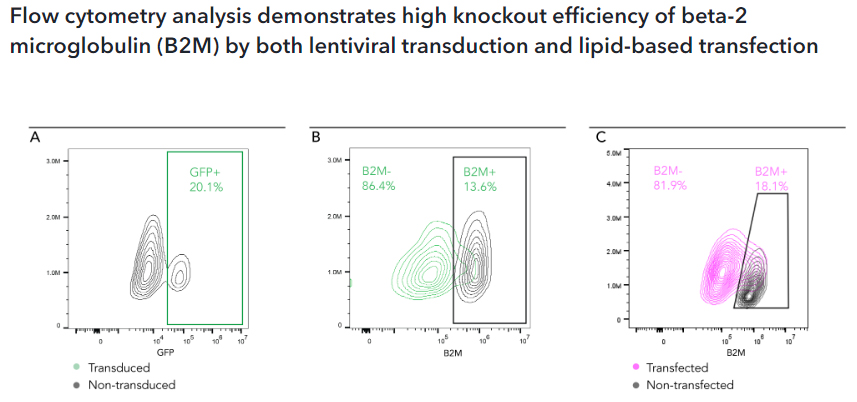
Flow cytometry analysis of B2M protein expression in CRISPR-Ready ioMicroglia, after delivery of gRNA targeting B2M. gRNAs were introduced into the cells at day 10 post-thaw using two methods: lentiviral transduction or synthetic gRNA delivery with Lipofectamine RNAiMAX transfection reagent. After 5 days of culture following guide delivery, B2M gene knockout efficiency was assessed by flow cytometry analysis. (A) Lentiviral transduction with gRNA targeting B2M: 20% of cells received a B2M gRNA, as measured by GFP expression. A high knockout efficiency of 86% was achieved in these GFP+ cells (B). (C) Lipid-based transfection with gRNA targeting B2M: a high knockout efficiency of 82% was achieved.
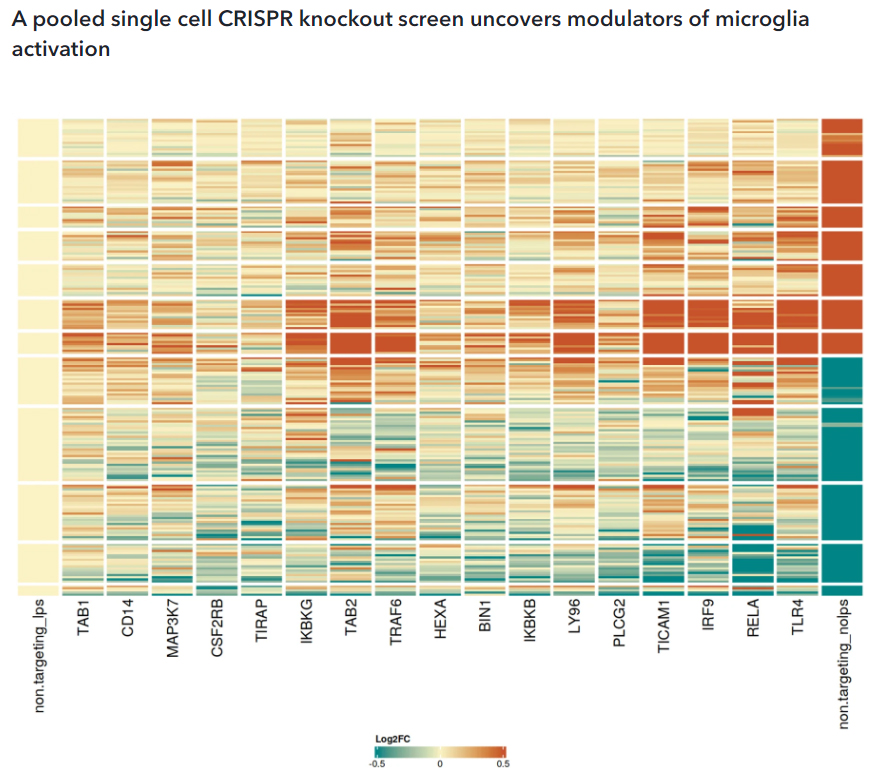
To conduct a pooled single cell CRISPR knockout screen (scCRISPR screen) with a targeted sequencing readout, we first identified a transcriptomic activation signature of 258 differentially expressed genes by comparing CRISPR-Ready ioMicroglia treated with and without LPS at day 10. Separately, we selected 110 candidate genes for the pooled scCRISPR screen based on their known roles in neurodegeneration and neuroinflammation. Guide RNAs were delivered via lentiviral transduction on day 10, aiming for a single integration per cell. The cells were cultured and then treated with +/- LPS for 24 hours before single cell processing on day 15. Cosine similarity analysis compared knockouts in LPS-treated CRISPR-Ready ioMicroglia to both resting and activated states. The analysis identified 17 gene knockouts that altered responses to LPS stimulation. The heatmap shows Log2FC profiles for gene knockouts that had a cosine similarity above 0.3 (arbitrarily chosen threshold) compared to cells with non-targeting guides in the unstimulated condition. Knockouts are sorted based on their cosine similarity to the non-LPS condition. CD14, MAP3K7, TIRAP, IKBKG, TRAF6, IKBKB, LY96, TICAM1, RELA, and TLR4 are genes known to be involved in LPS activation mediated via the TLR4 signalling pathway.
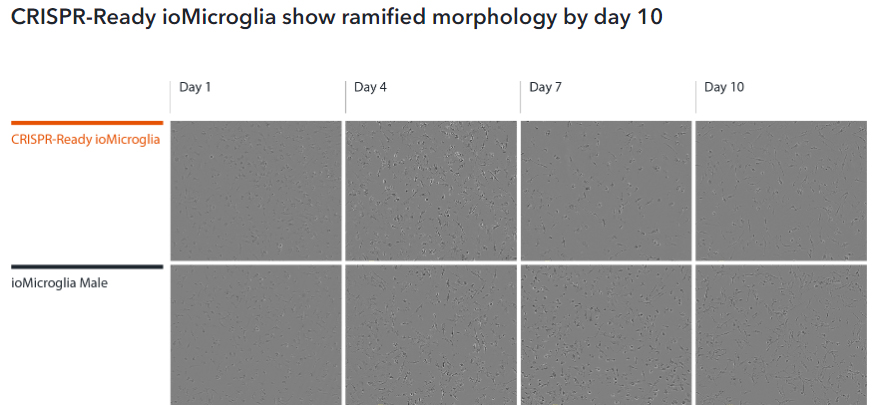
CRISPR-Ready ioMicroglia mature rapidly and key ramified morphology can be identified by day 4 and continues through to day 10, similarly to ioMicroglia Male (io1021). Day 1 to 10 post-thawing; 100x magnification.
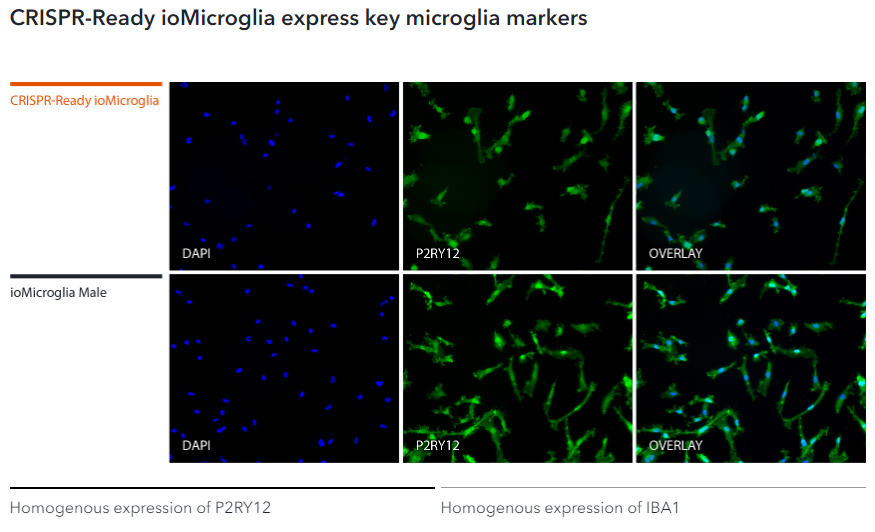
Immunofluorescent staining on day 10 post-revival demonstrates similar homogenous expression of microglia markers P2RY12 and IBA1 and ramified morphology in CRISPR-Ready ioMicroglia compared to ioMicroglia Male (io1021). 100X magnification.
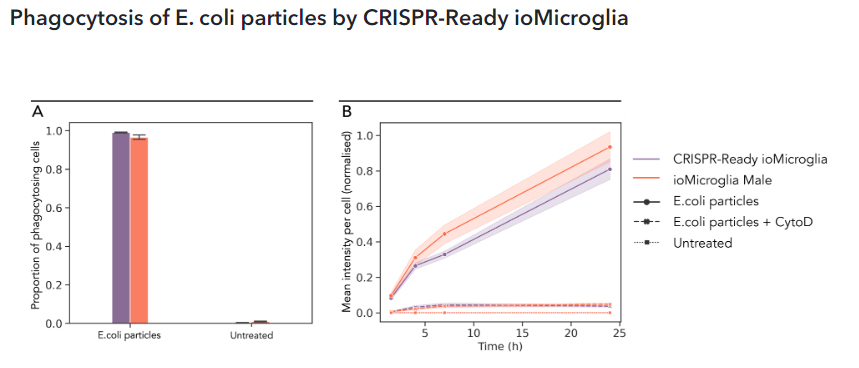
(A) Phagocytosis assay using pHrodo™ E. coli BioParticles™ at day 10 post-thaw demonstrates efficient uptake of bacteria particles by CRISPR-Ready ioMicroglia in comparison to ioMicroglia Male (io1021) after 4 hours of treatment. (B) An increase of fluorescence intensity of E.coli particles upon pH change in the phagosome, can be readily detected by fluorescence microscopy. A steep increase of fluorescence signal intensity was measured in the presence of E.coli particles alone, but not in combination with Cytochalasin D (CytoD), an inhibitor of actin polymerization.
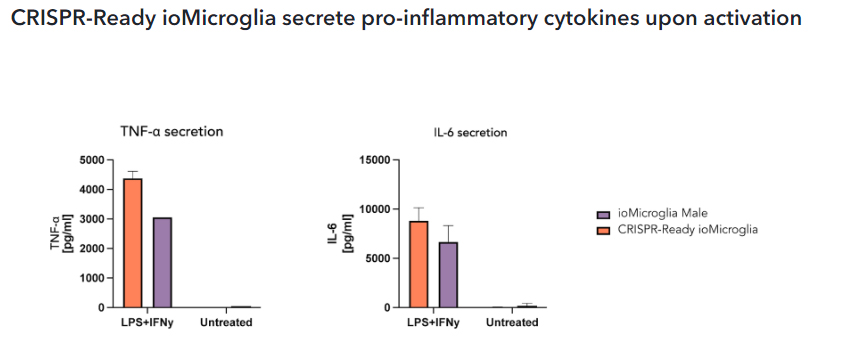
CRISPR-Ready ioMicroglia were stimulated at day 10 post-thaw with LPS 100 ng/ml and IFNɣ 20 ng/ml for 24 hours. Supernatants were harvested and analysed by quantitative ELISA. These cells secrete the pro-inflammatory cytokines, TNF⍺ and IL-6 upon activation in comparison to ioMicroglia Male (io1021).